Input Form
The user can paste in the text-area the protein sequences in FASTA format.
The maximum number of sequences that can be inserted is 100 when using only the sequence features.
The maximum lenght for each sequence is 5000 residues.
The user can choose to encode the sequences using one of the two following encodings:
- BINARY
- PAM30
The user can also choose between two combination of features:
- only the sequence around the site
- the sequence, the secondary structure and disorder prediction for the site and the composition features.
We suggest to try firstly to run the predictor using the binary encoding with only the sequence features, as the predictions are given in very short time and the performances are comparable to those obtained using all the features.
The prediction of secondary structure is time-consuming, therefore if the user choose the predictor with all the features the maximum limit of submitted sequences decreases to 5.
Note that if two sequences have the same identifier, the program stops and returns an error.
For more details about the features, please go to the Overview section
Output
The results are listed in a table, that contain the following fields, from left to right:
- the Accession of the sequence
- the sorrounding 11 residues around the site
- the position of the site in the sequence
- the PhosTryp score for the site.
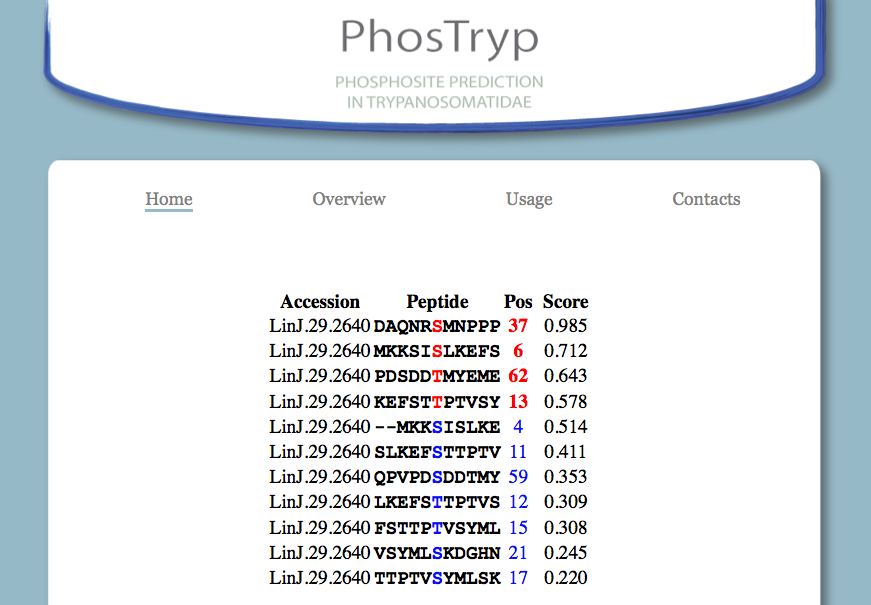
Phosphosites are marked in red. Sites with scores below the cutoff are in blue. By default all the sites are sorted by score.